Last Updated on 26/06/2022 by TheDigitalHacker
Artificial intelligence has helped to solve a 50-year-old riddle that could lead to faster virus treatments and medicinal discoveries.
Google-owned AI lab DeepMind, based in London, says its AlphaFold programme has solved the issue and is capable of predicting the structure that proteins will fold into.
The “protein folding problem” has baffled scientists for decades because there are so many different types of proteins and they can be found in all living organisms. Also, another major problem with proteins is the fact it’s very difficult to map out their 2D structures.
Coronavirus is related to the way proteins function as well as other diseases like dementia and cancer.
There are 200 million known proteins at present but only a fraction have actually been unfolded to fully understand what they do and how they work.
DeepMind has worked on the AI project with Scientists from the 14th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction (CASP14). They’ve been trying to solve the protein riddle since 1994.
What is Protein folding problem and How can AI make a difference?
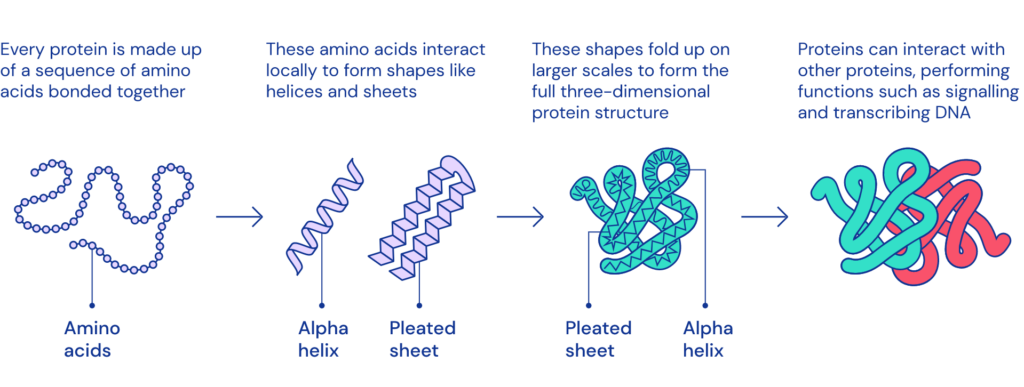
Proteins are large, complex molecules essential to all of life. Nearly every function that our body performs – contracting muscles, sensing light, or turning food into energy – relies on proteins, and how they move and change. What any given protein can do depends on its unique 3D structure.
Proteins are comprised of chains of amino acids (also referred to as amino acid residues). Listing a protein’s amino acids is easy. Machines to do so have existed for decades. But this is only half the battle in the quest to understand how proteins work.DNA only contains information about the sequence of amino acids – not how they fold into shape. What a protein does, and how it does it, depends also on how it folds up after its creation, into its final, intricate shape. The bigger the protein, the more difficult it is to model because there are more interactions between amino acids to take into account.
Predicting how these chains will fold into the intricate 3D structure of a protein is what’s known as the “protein folding problem” – a challenge that scientists have worked on for decades. This unsolved problem has already inspired countless developments, from spurring IBM’s efforts in supercomputing (BlueGene) to novel citizen science efforts to new engineering realms, such as rational protein design.
Scientists have long been interested in determining the structure of proteins because it is believed that the shape of a protein determines its function.
Once the form of the protein is understood, it will be possible to guess its role in the cell, and scientists will therefore be able to develop drugs that work with the unique form of the protein.
Over the past five decades, researchers have been able to determine the shapes of proteins in laboratories using experimental techniques such as cryo-electron microscopy, nuclear magnetic resonance, and x-ray crystallography. Each of these methods and many others is based on a lot of trial and error processes, which not only takes a lot of efforts and years of research but also tonnes of money for each protein structure.
This is why biologists are turning to artificial intelligence techniques as an alternative to this long and laborious process of processing complex proteins.
The ability to predict a protein’s shape using calculations based solely on its genetic code, rather than determining it through expensive experiments, could help speed the research.
Scientists say more work remains to be done and a paper containing their current research will have to be submitted for review. As we know, their discovery could lead to a change in scientific research.
DeepMind plans to publish a paper detailing AlphaFold’s achievement, but it was diffident on whether it would release the algorithm itself.
“We’re right at the beginning of exploring how best to enable other groups to use our structure predictions,” the company said.